github 仓库
https://github.com/chunleili/pbf3d
效果展示
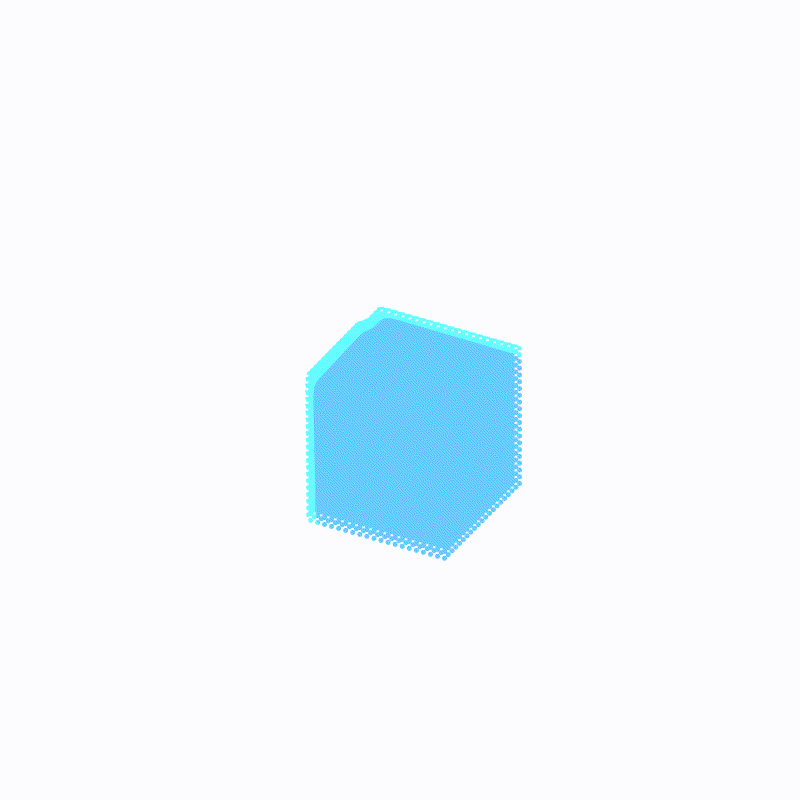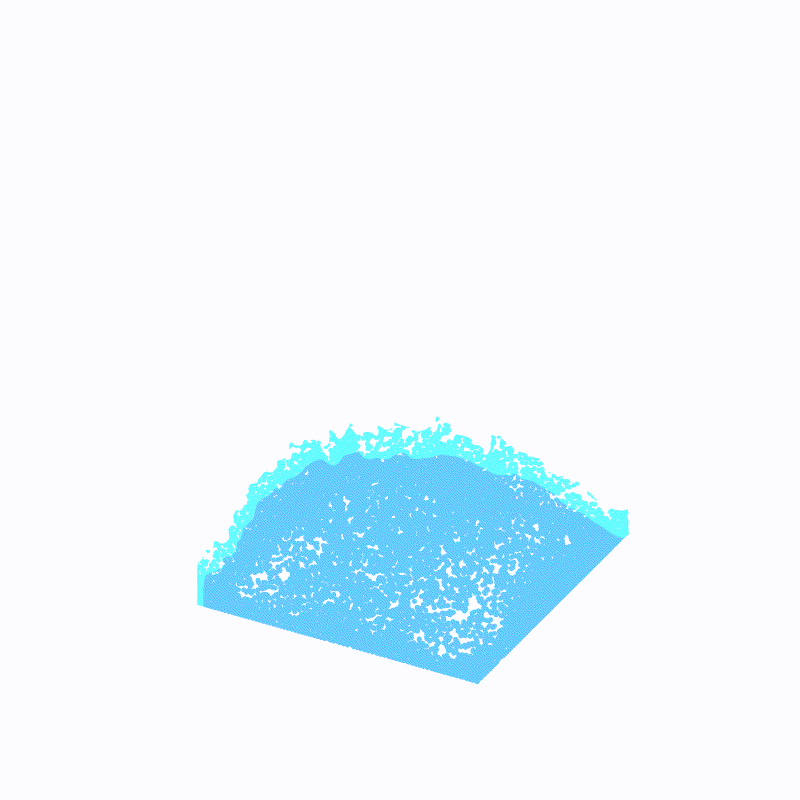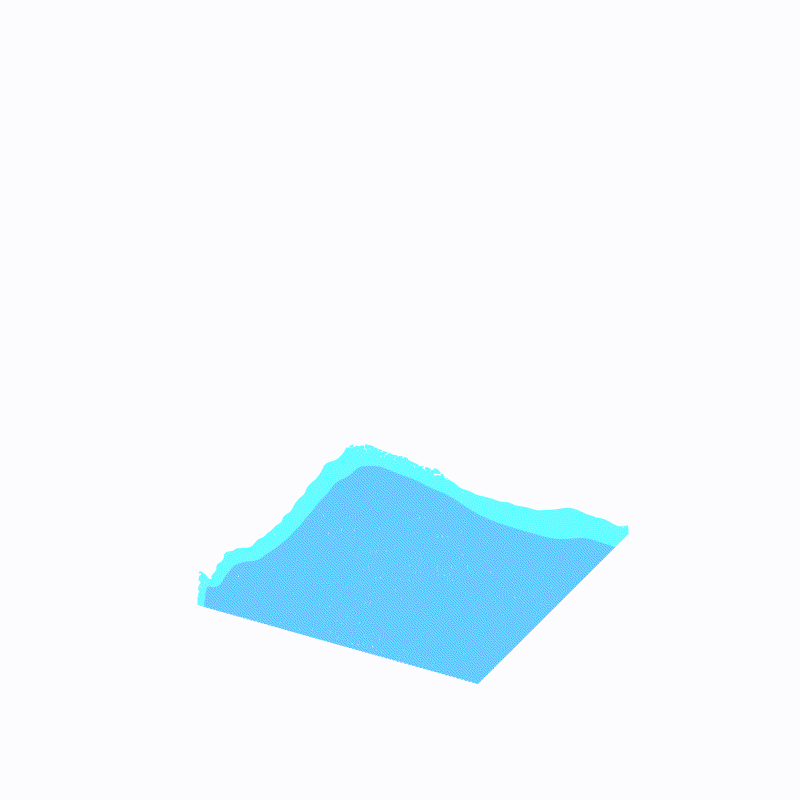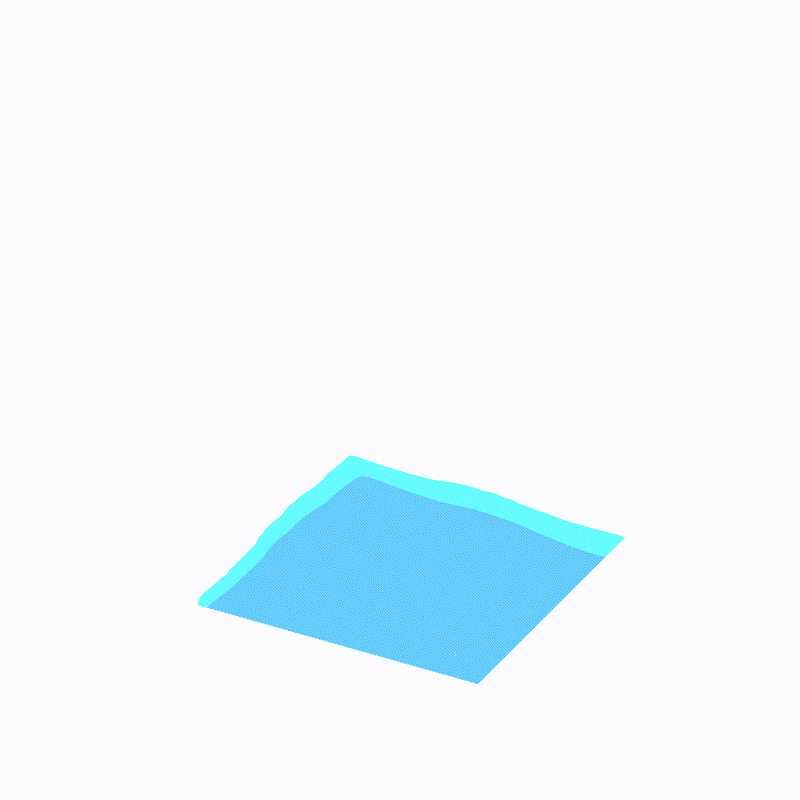
(最外层的浅蓝色是ffmpeg造成的,请忽略)
原理讲解
https://www.bilibili.com/video/BV1qa411X7Uo/
(我是小白,我讲的不对的地方请立即吐槽我)
PBF,基于位置流体的原始论文
- Macklin, M., Müller, M. 2013. Position Based Fluids. ACM Trans. Graph. 32, 4, Article 104, July 2013, 5 pages. http://doi.acm.org/10.1145/2461912.2461984
代码本身就写得很好,非常适合改造。更改的时候只需要注意以下几个要点:
避坑和技术总结如下
-
3d的,要么用GGUI(参考mpm3d_ggui那个例子),要么采用2d渲染3d(参考mpm3d那个例子)。所谓2d渲染3d其实就是自己做MVP变换(模型、视图、投影变换),这点直接复制mpm3d例子中的T函数即可把3d的粒子坐标投影到2d屏幕上。
-
所有出现ti.Vector([0.0, 0.0])的地方显然要改成ti.Vector([0.0, 0.0, 0.0])。同理ti.grouped(ti.ndrange((-1, 2), (-1, 2),(-1, 2)))这里改成3维度的。这都是因为太极ti.Vector等目前没法实现维度无关所造成的麻烦。
-
数据结构要注意grid_num_particles和grid2particles这两个数据结构。由于SNode暂时不能创建4维数组(因为grid_size本身是3维的,然后网格中的粒子数组是第四维度),所以干脆直接用ti.field指定shape的方式
grid_num_particles很简单,只要把ti.ij改为ti.ijk就行了
grid_snode = ti.root.dense(ti.ijk, grid_size)
grid_snode.place(grid_num_particles)
grid2particles = ti.field(int, (grid_size + (max_num_particles_per_cell,)))
这里要注意那个逗号。因为python当中小括号包裹的是元组,元组是可以拼接的,但是要先把int转换成元组,所以不得不加个逗号表示它不是一个数,而是一个元组。
-
初始排列那里要费一些小心思。但是也不是很难。参考我的上一个帖子。或者看这里http://t.csdn.cn/Kpku8。其实就是“排排坐,吃果果,一排排完再一排,一层排完再一层”。我没有直接修改k-ye的init_particles,而是干脆自己新写了一个init函数。
-
一些不必要的函数可以清理掉,比如move_board和相关的代码和变量。这个没啥用,只是加了个fancy的移动板子。
-
要注意调整世界大小。我这里指的是T函数里面最后return 原本+0.5, 我这里改成了25。为啥要改呢?因为mpm3d的世界大小是1.01.01.0的。而k-ye的代码世界大小是boundary(我们姑且认为boundary就是天空盒)。我这里改完了boundary的大小是(40,40,40)。你还可能发现我的gui.circles(T(pos)/100.0, radius=3, color=0x66ccff)这里除以了一百,这也是为了缩放世界的大小。
我的建议是:世界的大小遵循匡冶的设定,而不是mpm3d的设定。假如遵循mpm3d的设定,核半径h就会小于1,这会造成一个结果:在evaluate核函数的时候,会导致异常大的核函数值。因为在spiky kernel中h的6次方是要做分母的!
- 最后一个建议:修改代码时,保证的一个原则是最小化修改。所谓能不改的不改,不知道该不该改的不该,必须改的一口气改。要记住自己改了哪,没改哪。不要散弹枪式的修改。拿不准的函数或者特性就单独拎出来测试。谨慎使用自己还不是特别懂的新特性。(利用好git)
代码如下
# Macklin, M. and Müller, M., 2013. Position based fluids. ACM Transactions on Graphics (TOG), 32(4), p.104.
# Taichi implementation by Ye Kuang (k-ye)
# Modified by Chunlei Li, change to 3D, 2022/7/4
import math
import numpy as np
import taichi as ti
ti.init(arch=ti.gpu)
screen_res = (800, 800)
screen_to_world_ratio = 20.0
boundary = (screen_res[0] / screen_to_world_ratio,
screen_res[1] / screen_to_world_ratio,
screen_res[0] / screen_to_world_ratio,)
cell_size = 2.51
cell_recpr = 1.0 / cell_size
def round_up(f, s):
return (math.floor(f * cell_recpr / s) + 1) * s
grid_size = (round_up(boundary[0], 1), round_up(boundary[1], 1), round_up(boundary[2], 1))
dim = 3
bg_color = 0x112f41
particle_color = 0x068587
boundary_color = 0xebaca2
num_particles_x = 10
# num_particles = num_particles_x * num_particles_x * 10
num_particles = 10000
max_num_particles_per_cell = 100
max_num_neighbors = 100
time_delta = 1.0 / 20.0
epsilon = 1e-5
particle_radius = 3.0
particle_radius_in_world = particle_radius / screen_to_world_ratio
# PBF params
h = 1.1
mass = 1.0
rho0 = 1.0
lambda_epsilon = 100.0
pbf_num_iters = 5
corr_deltaQ_coeff = 0.3
corrK = 0.001
# Need ti.pow()
# corrN = 4.0
neighbor_radius = h * 1.05
poly6_factor = 315.0 / 64.0 / math.pi
spiky_grad_factor = -45.0 / math.pi
old_positions = ti.Vector.field(dim, float)
positions = ti.Vector.field(dim, float)
velocities = ti.Vector.field(dim, float)
grid_num_particles = ti.field(int)
# grid2particles = ti.field(int)
particle_num_neighbors = ti.field(int)
particle_neighbors = ti.field(int)
lambdas = ti.field(float)
position_deltas = ti.Vector.field(dim, float)
ti.root.dense(ti.i, num_particles).place(old_positions, positions, velocities)
grid_snode = ti.root.dense(ti.ijk, grid_size)
grid_snode.place(grid_num_particles)
# grid_snode.dense(ti.i, max_num_particles_per_cell).place(grid2particles) #this way cannot place a 4 dimension array
grid2particles = ti.field(int, (grid_size + (max_num_particles_per_cell,)))
nb_node = ti.root.dense(ti.i, num_particles)
nb_node.place(particle_num_neighbors)
nb_node.dense(ti.j, max_num_neighbors).place(particle_neighbors)
ti.root.dense(ti.i, num_particles).place(lambdas, position_deltas)
@ti.func
def poly6_value(s, h):
result = 0.0
if 0 < s and s < h:
x = (h * h - s * s) / (h * h * h)
result = poly6_factor * x * x * x
return result
@ti.func
def spiky_gradient(r, h):
result = ti.Vector([0.0, 0.0, 0.0])
r_len = r.norm()
if 0 < r_len and r_len < h:
x = (h - r_len) / (h * h * h)
g_factor = spiky_grad_factor * x * x
result = r * g_factor / r_len
return result
@ti.func
def compute_scorr(pos_ji):
# Eq (13)
x = poly6_value(pos_ji.norm(), h) / poly6_value(corr_deltaQ_coeff * h, h)
# pow(x, 4)
x = x * x
x = x * x
return (-corrK) * x
@ti.func
def get_cell(pos):
return int(pos * cell_recpr)
@ti.func
def is_in_grid(c):
# @c: Vector(i32)
return 0 <= c[0] and c[0] < grid_size[0] and 0 <= c[1] and c[
1] < grid_size[1]
@ti.func
def confine_position_to_boundary(p):
bmin = particle_radius_in_world
bmax = ti.Vector([boundary[0], boundary[1], boundary[2]
]) - particle_radius_in_world
for i in ti.static(range(dim)):
# Use randomness to prevent particles from sticking into each other after clamping
if p[i] <= bmin:
p[i] = bmin + epsilon * ti.random()
elif bmax[i] <= p[i]:
p[i] = bmax[i] - epsilon * ti.random()
return p
@ti.kernel
def prologue():
# save old positions
for i in positions:
old_positions[i] = positions[i]
# apply gravity within boundary
for i in positions:
g = ti.Vector([0.0, -9.8, 0.0])
pos, vel = positions[i], velocities[i]
vel += g * time_delta
pos += vel * time_delta
positions[i] = confine_position_to_boundary(pos)
# clear neighbor lookup table
for I in ti.grouped(grid_num_particles):
grid_num_particles[I] = 0
for I in ti.grouped(particle_neighbors):
particle_neighbors[I] = -1
# update grid
for p_i in positions:
cell = get_cell(positions[p_i])
# ti.Vector doesn't seem to support unpacking yet
# but we can directly use int Vectors as indices
offs = ti.atomic_add(grid_num_particles[cell], 1)
grid2particles[cell, offs] = p_i
# find particle neighbors
for p_i in positions:
pos_i = positions[p_i]
cell = get_cell(pos_i)
nb_i = 0
for offs in ti.static(ti.grouped(ti.ndrange((-1, 2), (-1, 2),(-1, 2)))):
cell_to_check = cell + offs
if is_in_grid(cell_to_check):
for j in range(grid_num_particles[cell_to_check]):
p_j = grid2particles[cell_to_check, j]
if nb_i < max_num_neighbors and p_j != p_i and (
pos_i - positions[p_j]).norm() < neighbor_radius:
particle_neighbors[p_i, nb_i] = p_j
nb_i += 1
particle_num_neighbors[p_i] = nb_i
@ti.kernel
def substep():
# compute lambdas
# Eq (8) ~ (11)
for p_i in positions:
pos_i = positions[p_i]
grad_i = ti.Vector([0.0, 0.0, 0.0])
sum_gradient_sqr = 0.0
density_constraint = 0.0
for j in range(particle_num_neighbors[p_i]):
p_j = particle_neighbors[p_i, j]
if p_j < 0:
break
pos_ji = pos_i - positions[p_j]
grad_j = spiky_gradient(pos_ji, h)
grad_i += grad_j
sum_gradient_sqr += grad_j.dot(grad_j)
# Eq(2)
density_constraint += poly6_value(pos_ji.norm(), h)
# Eq(1)
density_constraint = (mass * density_constraint / rho0) - 1.0
sum_gradient_sqr += grad_i.dot(grad_i)
lambdas[p_i] = (-density_constraint) / (sum_gradient_sqr +
lambda_epsilon)
# compute position deltas
# Eq(12), (14)
for p_i in positions:
pos_i = positions[p_i]
lambda_i = lambdas[p_i]
pos_delta_i = ti.Vector([0.0, 0.0, 0.0])
for j in range(particle_num_neighbors[p_i]):
p_j = particle_neighbors[p_i, j]
if p_j < 0:
break
lambda_j = lambdas[p_j]
pos_ji = pos_i - positions[p_j]
scorr_ij = compute_scorr(pos_ji)
pos_delta_i += (lambda_i + lambda_j + scorr_ij) * \
spiky_gradient(pos_ji, h)
pos_delta_i /= rho0
position_deltas[p_i] = pos_delta_i
# apply position deltas
for i in positions:
positions[i] += position_deltas[i]
@ti.kernel
def epilogue():
# confine to boundary
for i in positions:
pos = positions[i]
positions[i] = confine_position_to_boundary(pos)
# update velocities
for i in positions:
velocities[i] = (positions[i] - old_positions[i]) / time_delta
# no vorticity/xsph because we cannot do cross product in 2D...
def run_pbf():
prologue()
for _ in range(pbf_num_iters):
substep()
epilogue()
@ti.kernel
def init():
init_pos = ti.Vector([10.0,10.0,10.0])
cube_size = 20
spacing = 1
num_per_row = (int) (cube_size // spacing) + 1
num_per_floor = num_per_row * num_per_row
for i in range(num_particles):
floor = i // (num_per_floor)
row = (i % num_per_floor) // num_per_row
col = (i % num_per_floor) % num_per_row
positions[i] = ti.Vector([col*spacing, floor*spacing, row*spacing]) + init_pos
def T(a):
if dim == 2:
return a
phi, theta = np.radians(28), np.radians(32)
a = a - 0.5
x, y, z = a[:, 0], a[:, 1], a[:, 2]
c, s = np.cos(phi), np.sin(phi)
C, S = np.cos(theta), np.sin(theta)
x, z = x * c + z * s, z * c - x * s
u, v = x, y * C + z * S
return np.array([u, v]).swapaxes(0, 1) + 25
def main():
init()
# init_particles()
print(f'boundary={boundary} grid={grid_size} cell_size={cell_size}')
gui = ti.GUI('PBF3D', screen_res, background_color = 0xffffff)
while gui.running and not gui.get_event(gui.ESCAPE):
run_pbf()
pos = positions.to_numpy()
# print(pos)
export_file = ""
if export_file:
writer = ti.tools.PLYWriter(num_vertices=num_particles)
writer.add_vertex_pos(pos[:, 0], pos[:, 1], pos[:, 2])
writer.export_frame(gui.frame, export_file)
gui.circles(T(pos)/100.0, radius=3, color=0x66ccff)
gui.show()
if __name__ == '__main__':
main()