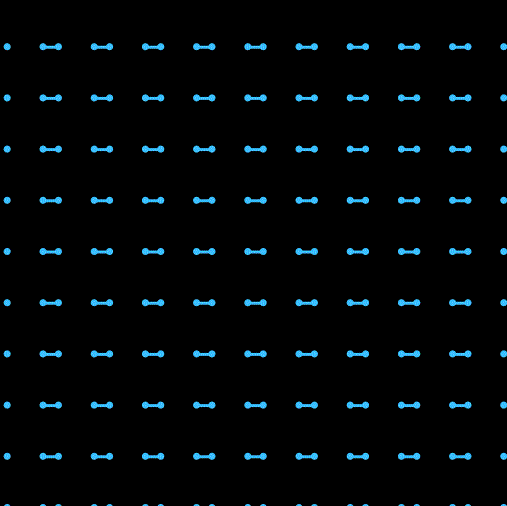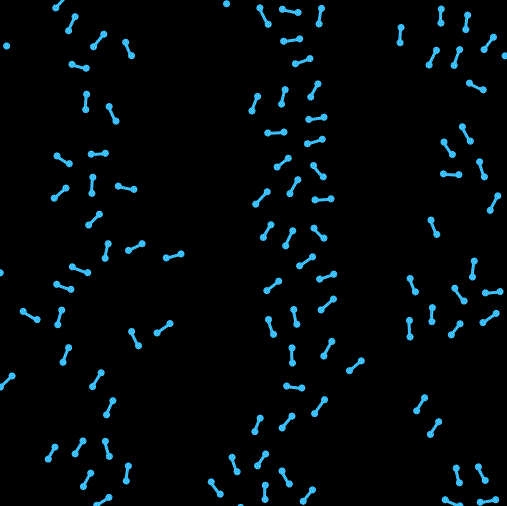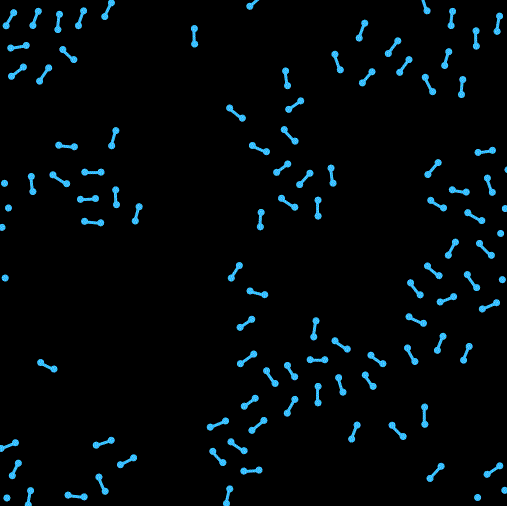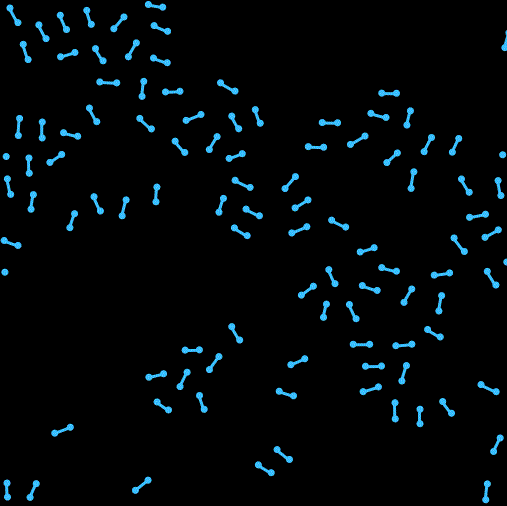
Hi我又来了。这次在上一次单原子的基础上尝试写了一个双原子的模拟。之前一直头大该如何处理分子内两个原子位置和取向之类的问题。最开始想尝试写一个完全刚体的分子,用作用在两个原子上的合力来确定质心的移动,然后用力矩来确定分子取向的转动,结果没写好 ![]() 分子的转向会出现爆炸等情况(不知道用隐式时间积分能不能行,还是本身写的有问题)。最后换成了两个分子间用硬弹簧链接的形式来暂时解决了这个问题,另外还加入了周期性边界条件。
分子的转向会出现爆炸等情况(不知道用隐式时间积分能不能行,还是本身写的有问题)。最后换成了两个分子间用硬弹簧链接的形式来暂时解决了这个问题,另外还加入了周期性边界条件。
代码如下:
from taichi_glsl import *
import numpy as np
ti.init()
max_num_pair = 512
particle_mass = 1.0
bond_length = 0.03
# I = particle_mass * half_bond_length ** 2 # Momentum of Inertia
dt = 1e-3
substeps = 10
# Force parameter
spring_Y = 1000
sigma = 0.04
sigma2 = sigma ** 2
sigma6 = sigma2 ** 3
sigma12 = sigma6 ** 2
potential_energy = 0.1
cutoff_r = 0.2
paused = ti.field(dtype=ti.i32, shape=())
x = ti.Vector.field(2, dtype=ti.f32, shape=(max_num_pair,2))
v = ti.Vector.field(2, dtype=ti.f32, shape=(max_num_pair,2))
f = ti.Vector.field(2, dtype=ti.f32, shape=(max_num_pair,2))
num_pairs = ti.field(dtype=ti.i32, shape=())
@ti.kernel
def set_force():
n = num_pairs[None]
for i in range(0, n):
for j in range(i + 1, n):
for a in range(2):
for b in range(2):
x_ab = x[i,a] - x[j,b]
x_ab -= round(x_ab[0])
if x_ab.norm() <= cutoff_r:
delta_x = x_ab.norm()
x2 = delta_x ** 2
x6 = x2 ** 3
x7 = x6 * delta_x
x13 = x6 ** 2 * delta_x
d = x_ab.normalized()
f[i, a] += (24 * potential_energy * (2 * sigma12 / x13 - sigma6 / x7)) * d
f[j, b] -= (24 * potential_energy * (2 * sigma12 / x13 - sigma6 / x7)) * d
@ti.kernel
def set_v():
n = num_pairs[None]
for i in range(n):
rab = x[i,1] - x[i,0]
rab -= round(rab)
d = rab.normalized()
fab = -spring_Y * (rab.norm()/bond_length - 1) * d
f[i,0] += -fab
f[i,1] += fab
for a in range(2):
v[i,a] += dt * f[i,a]/particle_mass
x[i,a] += dt * v[i,a]
x[i, a] -= round(x[i,a]) - 0.5
f[i,a] = ti.Vector([0, 0])
@ti.kernel
def add_pair(x_pos: ti.f32, y_pos: ti.f32, theta: ti.f32):
new_pair_id = num_pairs[None]
dx = ti.Vector([cos(theta), sin(theta)]) * bond_length/2
x[new_pair_id,0] = ti.Vector([x_pos, y_pos]) + dx
x[new_pair_id,1] = ti.Vector([x_pos, y_pos]) - dx
num_pairs[None] += 1
def main():
gui = ti.GUI("Diatomic Molecule",
res=512,
background_color=0x0)
# create pairs
for i in range(10):
for j in range(10):
add_pair(i/10, j/10, 0)
while gui.running:
for substep in range(substeps):
set_force()
set_v()
X = x.to_numpy()
n = num_pairs[None]
c = 0x3CC1FF
for i in range(n):
for a in range(2):
gui.circle(X[i,a], color=c, radius=4)
if np.linalg.norm(X[i,0]-X[i,1]) < 0.5:
gui.line(begin=X[i,0],end=X[i,1],radius=2,color=c)
gui.show()
if __name__ == '__main__':
main()
以及运行效果: